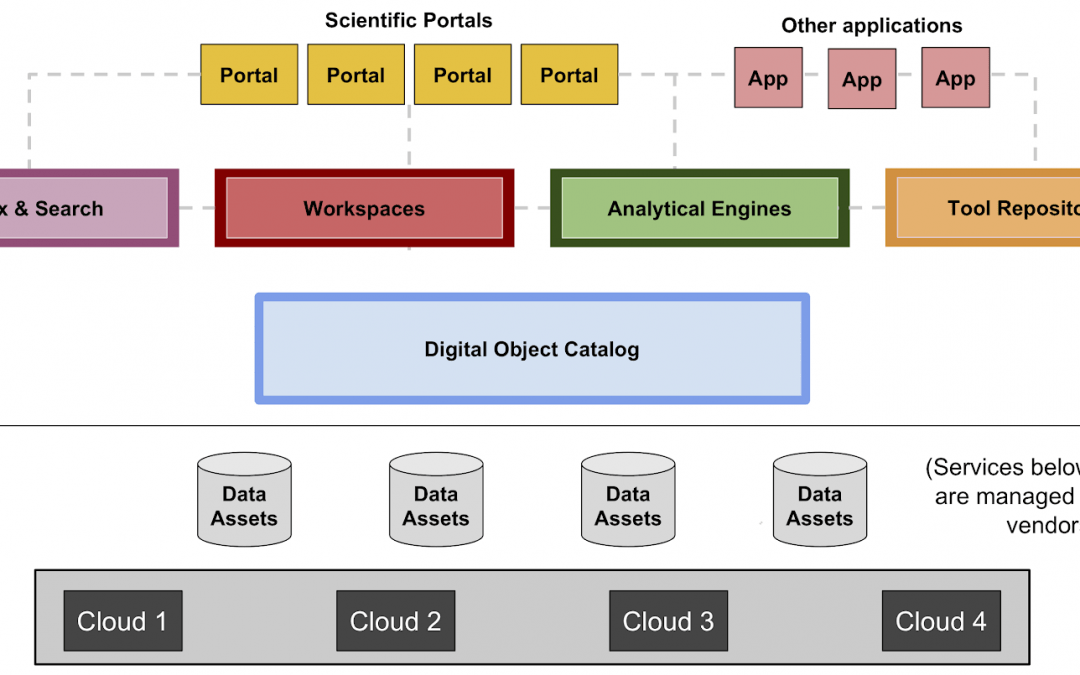
In 2017, Benedict Paten and collaborators proposed the “Data Biosphere” as a means to substantially accelerate genomics and biomedical research by building cloud platforms where components are modular, open and based on community standards (such as those developed by the Global Alliance for Genomics and Health (GA4GH)). Each platform contains portals for secure access to data, repositories for genomics tools and workflows, and cloud based compute platforms. This inversion of the traditional data sharing model brings researchers directly to the data and removes the burden of having to download and store the petabytes of available data.
Fast forward to today, and the Computational Genomics Platform (CGP) team at the UC Santa Cruz Genomics Institute is actively working on delivery of multiple platforms that each hold rich genomic datasets that are highlighted below. The ultimate goal for these platforms is interoperability, creating the opportunity for researchers to combine and analyze multi-omic datasets from various sources in a secure and scalable cloud infrastructure.
- Dockstore is a collaboration between UCSC and Canada’s Ontario Institute of Cancer Research, and is becoming the standard for the sharing of Docker-based workflows and tools for genomic analysis. Dockstore interoperates with multiple data biosphere workspaces, including Terra and Seven Bridges.
- BioData Catalyst is the NHLBI platform focused on providing access to over 150,000 whole genome sequences from sources including TOPmed using the Gen3 service and integrating with secure compute infrastructure like Terra and Seven Bridges.
- The AnVIL is the NHGRI platform providing access to multi-omic data sources such as CCDG, CMG, eMERGE, and the Human Pangenome Project using the Gen3 data repository and Terra compute infrastructure.
- The Human Cell Atlas provides access to community generated, multi-omic, open-access data, allowing researchers to understand what is unique to each cell in the human body.
These platforms are currently in early releases and they continue to evolve, but already the aspirational goals of that 2017 article are providing practical capabilities to researchers across the globe to increase their access to data, tools and compute and thereby accelerating greatly our learning about the true nature of the human genome.To learn more or to get involved in these platforms, please contact Tim Harris or Beth Sheets at the UC Santa Cruz Genomic Institute.
Recent Comments